
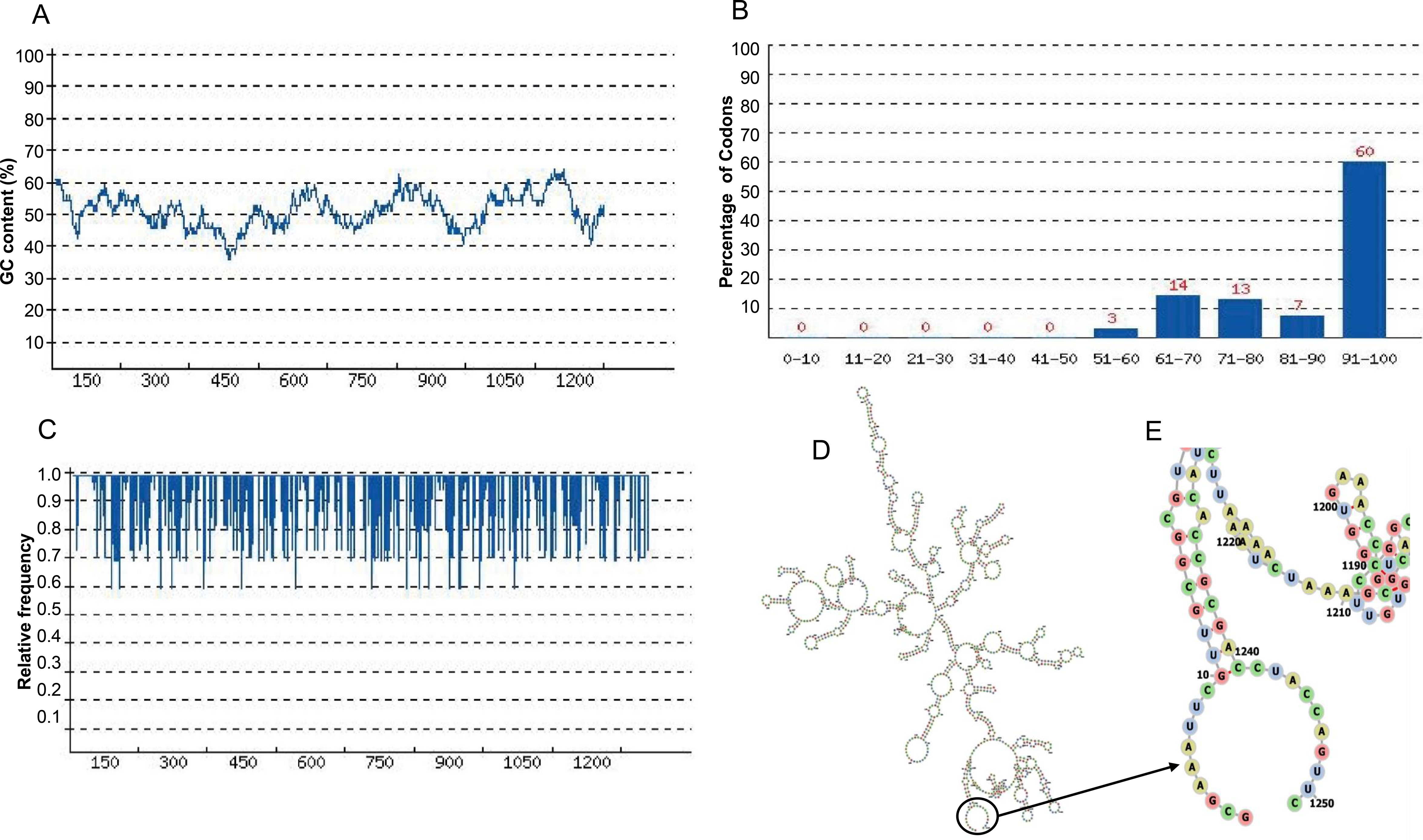
Although codon optimization has been shown to alter polysome profiles of some mRNAs ( 8, 14), which can be used as an indication of translation efficiency, the interpretation is complicated by the fact that codon usage also directly influences ribosome decoding rate ( 15–17). In addition, genome-wide correlations between protein levels and codon usage have been observed ( 12, 13). Selection for efficient translation is thought to be the major cause of codon usage bias ( 4, 9–11) however, the molecular mechanism that mediates the codon usage influence on mRNA translation efficiency is unclear.Ĭodon optimization has long been used to enhance heterologous gene expression, and highly expressed proteins are mostly encoded by genes with strong codon biases for optimal codons. Codon usage bias is an important determinant of gene expression levels in both eukaryotes and prokaryotes ( 5–8). Preference for certain synonymous codons, a phenomenon called codon usage bias, is a universal feature of all genomes ( 1–4). Most amino acids are encoded by two to six synonymous codons. Silencing of eRF1 expression resulted in codon usage-dependent changes in protein expression. Together, these results establish a mechanism for how codon usage regulates mRNA translation efficiency. In both Neurospora and Drosophila cells, codon usage plays an important role in regulating mRNA translation efficiency. We found that the rare codon-dependent premature termination is mediated by the translation termination factor eRF1, which recognizes ribosomes stalled on rare sense codons. This mechanism was shown to be conserved in Drosophila cells. Rare codons resulted in ribosome stalling in manners both dependent and independent of protein sequence context and caused premature translation termination. By analyzing ribosome profiling results, here we showed that codon usage regulates translation elongation rate and that rare codons are decoded more slowly than common codons in all codon families in Neurospora. A major role of codon usage is thought to regulate protein expression levels by affecting mRNA translation efficiency, but the underlying mechanism is unclear. Codon usage bias is a universal feature of eukaryotic and prokaryotic genomes and plays an important role in regulating gene expression levels.


 0 kommentar(er)
0 kommentar(er)
